When the anomaly data is very few, we need to make good use of F1 Score to help us to figure out which model is better.
Gaussian Distribution
plot the data
# import modules
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
import scipy.io # used to load the mat files
import scipy.optimize
# load the data into numpy
filename = 'ex8/data/ex8data1.mat'
mat = scipy.io.loadmat(filename)
print mat['X'].shape,mat['Xval'].shape,mat['yval'].shape
(307, 2) (307, 2) (307, 1)
X = mat['X']
Xval = mat['Xval']
yval = mat['yval']
def plotData(myX, newFig = True):
if newFig:
plt.figure(figsize=(8,6))
plt.plot(myX[:,0],myX[:,1],'b+')
plt.xlabel('Latency [ms]',fontsize=16)
plt.ylabel('Throughput [mb/s]',fontsize=16)
plt.grid(True)
plt.show()
plotData(X)

cal the mean and deviration
def estimateGaussian(myX):
"""
compute the mean and deviration of the array-like:myX, then return the mean and deviration
"""
mu = np.mean(myX,axis=0)
deviration = myX - mu
dev2 = np.sum(np.power(deviration,2),axis=0)
return mu,dev2 / myX.shape[0]
mu,dev2 = estimateGaussian(X)
print mu.shape, dev2
(2,) [ 1.83263141 1.70974533]
cal the probability
print np.diag(dev2),np.linalg.det(np.diag(dev2))
the output is:
[[ 1.83263141 0. ]
[ 0. 1.70974533]]
3.13333300235
def gaus(myX, mymu, mysig2):
"""
Function to compute the gaussian return values for a feature,
matrix, myX, given the already computed mu vector and sigma matrix.,
If sigma is a vector, it is turned into a diagonal matrix,
Uses a loop over rows; I didn't quite figure out a vectorized implementation.
"""
m = myX.shape[0]
n = myX.shape[1]
if np.ndim(mysig2) == 1:
mysig2 = np.diag(mysig2)
norm = 1.0 / (np.power(2.0 * np.pi, n /2.0) * np.sqrt(np.linalg.det(mysig2)))
myinv = np.linalg.inv(mysig2)
myexp = np.zeros((m,1))
for irow in xrange(m):
xrow= myX[irow]
myexp[irow] = np.exp(-0.5*((xrow-mymu).T).dot(myinv).dot(xrow-mymu))
return norm * myexp
p = gaus(X,mu,dev2)
print p[:10]
the output is:
[[ 0.06470829]
[ 0.05030417]
[ 0.07245035]
[ 0.05031575]
[ 0.06368497]
[ 0.04245832]
[ 0.04790945]
[ 0.03651115]
[ 0.0186658 ]
[ 0.05068826]]
contour the gaussian distribution
def plotContours(myX, mymu, mysigma2, newFig=False, useMultivariate = True):
delta = 0.5
myx = np.arange(0,30,delta)
myy = np.arange(0,30,delta)
meshx,meshy = np.meshgrid(myx,myy)
coord_list = [ entry.ravel() for entry in (meshx, meshy) ]
points = np.vstack(coord_list)
myz = gaus(points.T,mymu,mysigma2)
#print myz[:,0].shape
myz = myz.reshape((myx.shape[0],myx.shape[0]))
if newFig:
plt.figure(figsize=(8,6))
plt.plot(myX[:,0],myX[:,1],'b+')
plt.xlabel('Latency [ms]',fontsize=16)
plt.ylabel('Throughput [mb/s]',fontsize=16)
plt.grid(True)
cont_levels = [10**exp for exp in range(-20,0,3)]
mycont = plt.contour(meshx,meshy,myz,cont_levels)
plt.title('Gaussian Contours',fontsize=16)
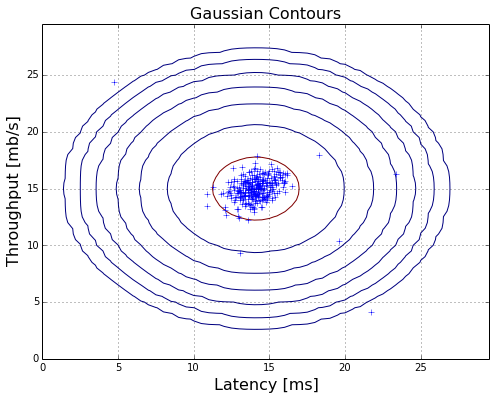
plotContours(X,mu,dev2,True,True)
#plt.show()
F1 score
implementation
def selectThreshold(yval,pval):
"""
yval:label
pval:predict value
function: according to both yval and pval to cal the F1 Score
"""
yval = yval.flatten()
pval = pval.flatten()
bestEpis = 0.0
bestF1 = 0.0
m = len(yval)
step = (np.max(pval) - np.min(pval)) / float(1000)
minv = np.min(pval)
for v in xrange(1000):
episode = minv + v * step
tp = np.sum(np.bitwise_and(pval < episode, yval==1))
fp = np.sum(np.bitwise_and(pval < episode, yval==0))
fn = np.sum(np.bitwise_and(pval > episode, yval==1))
#print tp,fp,fn
if tp + fp ==0:
continue
pre = float(tp)/(tp + fp)
if tp + fn == 0:
continue
rec = float(tp)/(tp + fn)
if (pre + rec)==0:
continue
F1 = 2 * pre * rec/(pre + rec)
if F1 > bestF1:
bestF1 = F1
bestEpis = episode
return bestF1, bestEpis
pval = gaus(Xval,mu,dev2)
bestF1, bestEpis = selectThreshold(yval,pval)
print "best F1 score is:%s,and best episode is:%s)" % ( bestF1, bestEpis )
best F1 score is:0.875,and best episode is:8.99085277927e-05)
visualization
def plotAnomalies(myX,mybestEps, newFig = False):
ps = gaus(myX,*estimateGaussian(myX))
ps = ps.flatten()
anoms = myX[ps < mybestEps]
if newFig:
plt.figure(figsize=(6,4))
plt.scatter(anoms[:,0],anoms[:,1],s=80,facecolors='none',edgecolors='r')
plotContours(X,mu,dev2,True,True)
plotAnomalies(X,bestEpis,False)
#plt.show()

test on 11 dimension samples
filename = 'ex8/data/ex8data2.mat'
mat = scipy.io.loadmat(filename)
X=mat['X']
Xval = mat['Xval']
yval = mat['yval']
print X.shape,Xval.shape,yval.shape
(1000, 11) (100, 11) (100, 1)
mu,sigma2 = estimateGaussian(X)
p = gaus(X,mu,sigma2)
pval = gaus(Xval,mu,sigma2)
bestF1, bestEpis = selectThreshold(yval,pval)
print 'Best epsilon found using cross-validation:%s', bestEpis
print 'Best F1 found on cross validation set: %s', bestF1
print '#Outliers found: %d' % np.sum(p < bestEpis)
Best epsilon found using cross-validation:%s 1.37722889076e-18 Best F1 found on cross validation set: %s 0.615384615385 #Outliers found: 117