2 main algorithms to help compress:
- K-Means Clustering
- Principal Component Analysis(PCA)
1. K-Means Clustering
1.1 import modules
#show the figures buildin the notebook
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
import scipy.io #Used to load the OCTAVE *.mat files
import scipy.misc #Used to show matrix as an image
from scipy import optimize
import random
1.2 import sample data
fileName = 'ex7/data/ex7data2.mat'
mat = scipy.io.loadmat(fileName)
X = mat['X']
1.3 find the closest for each sample
the algorithm as the following:
for i in xrange(iteration):
idx = findClosedCentroids(X,centroids)
centroids = computeMeans(X, idx, K)
def findClosedCentroid(X, centroids):
"""
return the indexs of the centroids for each sample in X
"""
ids = np.zeros((X.shape[0],1)) * (-1)
for i in xrange(X.shape[0]):
ids[i] = findClosest(X[i],centroids)
return ids
def findClosest(point,centroids):
"""
find the closest one in centroids array for the given sample
return the index of the closed one
"""
result = np.zeros((len(centroids),1))
for i in xrange(len(centroids)):
result[i] = distSquared(point, centroids[i])
return np.argmin(result,axis=0)
def distSquared(point1, point2):
"""
return the distance of these 2 points
"""
assert point1.shape == point2.shape
return np.sum(np.square(point1 - point2))
def computeCentroids(X, ids):
"""
first, according the distinguish values in ids to get the value:K
then, make good use of K to compute the K-centroids points(mean value)
"""
k = len(np.unique(ids))
result = np.empty((k,X.shape[1]))
#print result.shape
for i in xrange(k):
result[i]=np.mean(X[ids.flatten()==i],axis=0)
return result
initial_centroids = np.array([[3,3],[6,2],[8,5]]);
ids = findClosedCentroid(X,initial_centroids)
centroids = computeCentroids(X,ids)
print "the index for the init centroids:%s" % ids[0:3].flatten()
print "the first centroids we compute by the init centroids is:%s" % centroids
the output is:
the index for the init centroids:[ 0. 2. 1.] the first centroids we compute by the init centroids is:[[ 2.42830111 3.15792418] [ 5.81350331 2.63365645] [ 7.11938687 3.6166844 ]]
def calMeanK(X, centroids, maxIter=100):
cent_history=[]
for i in xrange(maxIter):
cent_history.append(centroids)
ids = findClosedCentroid(X,centroids)
cent = computeCentroids(X,ids)
if np.sum(cent!=centroids) == 0:
break
else:
centroids = cent
cent_history.append(cent)
return ids,cent_history
ids,cent_history=calMeanK(X,initial_centroids,100)
print cent_history
The output is:
[array([[3, 3],
[6, 2],
[8, 5]]), array([[ 2.42830111, 3.15792418],
[ 5.81350331, 2.63365645],
[ 7.11938687, 3.6166844 ]]), array([[ 2.31325526, 3.22830617],
[ 5.33273768, 2.43159599],
[ 6.8653618 , 3.23293995]]), array([[ 2.19692479, 3.42136707],
[ 4.83555397, 2.12976745],
[ 6.6560054 , 3.0751355 ]]), array([[ 1.98241171, 4.0250785 ],
[ 3.91150763, 1.47060546],
[ 6.34008592, 3.05366642]]), array([[ 1.95399466, 5.02557006],
[ 3.12663743, 1.1121712 ],
[ 6.12919526, 3.01606258]]), array([[ 1.95399466, 5.02557006],
[ 3.04367119, 1.01541041],
[ 6.03366736, 3.00052511]]), array([[ 1.95399466, 5.02557006],
[ 3.04367119, 1.01541041],
[ 6.03366736, 3.00052511]])]
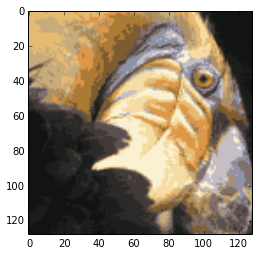
compress the image
load and display
fileName = 'ex7/data/bird_small.png'
A = scipy.misc.imread(fileName)
print "the loaded image's data shape:",A.shape
plt.figure()
dummy = plt.imshow(A)
plt.show()
the output is:
the loaded image’s data shape: (128, 128, 3)

normalization
A = A / 255.0 # after the normalization, then the feature is in the range[0,1]
print A.shape
B = A.reshape((A.shape[0] * A.shape[1],3)) # can also use (-1,3)
myK = 16 # use the 16 centroid point to stand for all the points in the image
init_centroids = random.sample(B, myK)
print init_centroids
the output is:
(128, 128, 3)
[array([ 0.47058824, 0.31764706, 0.21568627]), array([ 0.89803922, 0.76470588, 0.4745098 ]), array([ 0.57254902, 0.50196078, 0.17647059]), array([ 0.09411765, 0.10980392, 0.12156863]), array([ 0.06666667, 0.07058824, 0.05882353]), array([ 0.84313725, 0.69019608, 0.39215686]), array([ 0.69019608, 0.54901961, 0.56862745]), array([ 0.36078431, 0.22745098, 0.21960784]), array([ 0.43921569, 0.3254902 , 0.33333333]), array([ 0.23529412, 0.21960784, 0.21568627]), array([ 0.76470588, 0.6745098 , 0.41176471]), array([ 0.99607843, 0.95294118, 0.74509804]), array([ 0.11764706, 0.1254902 , 0.1254902 ]), array([ 0.85882353, 0.76078431, 0.52941176]), array([ 0.92156863, 0.86666667, 0.5254902 ]), array([ 0.82745098, 0.70196078, 0.45490196])]
cal the centroids
idxs, centroid_history = calMeanK(B,init_centroids,10)
print idxs
[[ 1.]
[ 1.]
[ 1.]
...,
[ 9.]
[ 12.]
[ 12.]]
compress and display
final_cent = centroid_history[-1]
cover_A = np.array([final_cent[int(i)] for i in idxs.flatten()])
cover_B = cover_A.reshape(A.shape)
plt.figure()
plt.imshow(cover_B)
plt.show()